bioinfo.pte.hu
bioinfo.pte.hu
BIOinformatika
Jegyzet a PTE TTK "Bioinformatika" c. biológia BSc. PTE TTK Genetikai és Molekuláris Biológiai Tanszék. Bevezető - 2000. Bevezető - 2003. Fehérje domének és más. Szekvencia javítás, illesztés. Régebbi, már nem gondozott fejezetek:. 1 Informatika és internet. 2 Bioinformatika és internet (lásd fent). 3 Minimális UNIX ismeretek. 4 A GCG programcsomag. 5 Az EMBOSS programcsomag. 6 A DNS 50 éve - cikkek, linkek. 7 DNS, gének, fehérjék. Számos prokariota genom, az élesztő ( Saccharomyces cerevisiae. A bioinfo...
 bioinfo.ru
bioinfo.ru
bioinfo.ru
This domain is for sale! Got questions about buying this domain? Call us and have the answers:. The domain is actually the site name and its correct choice influences greatly on the your endeavors success. Often, cost of the domain change during the project running is comparable with the re-branding, so the best way is to choose the right domain at the beginning, and to position your product or service once precisely. By submitting this form, you agree to the Terms.
 bioinfo.sastra.edu
bioinfo.sastra.edu
Zimbra Web Client Sign In
Offers the full set of Web collaboration features. This Web Client works best with newer browsers and faster Internet connections. Is recommended when Internet connections are slow, when using older browsers, or for easier accessibility. Is recommended for mobile devices. To be your preferred client type, change the sign in options in your Preferences, General tab after you sign in. Go offline with Zimbra Desktop. Learn more. The leader in open source messaging and collaboration : Blog.
 bioinfo.science.cmu.ac.th
bioinfo.science.cmu.ac.th
Untitled Document
ข าพเจ าม ความย นด ท ได มาร วมในพ ธ เป ดมหาว ทยาล ยเช ยงใหม ในว นน และม ความพอใจมากท ได ทราบความประสงค ในการจ ดต ง ตลอดถ งประโยชน ต าง ๆ อ นจะได ร บจากการต งมหาว ทยาล ยแห งน. บ ดน ถ งเวลากำหนดแล ว ข าพเจ าขอเป ดมหาว ทยาล ยเช ยงใหม ขอให สถานศ กษาแห งน ม ความเจร ญย งย นเป นประโยชน แก การศ กษาของก ลบ ตร ก ลธ ดา และแก ประเทศชาต โดยส วนรวมส บไปสมตามว ตถ ประสงค ท กประการ ขอให ท กท านท ประช มพร อมก นอย ณ ท น จงม ความส ขความเจร ญท วก น. ขอจงทรงพระเจร ญ ด วยเกล าด วยกระหม อม ขอเดชะ.
 bioinfo.sdu.edu.cn
bioinfo.sdu.edu.cn
生物信息学
请点击 https:/ vpn.sdu.edu.cn.
 bioinfo.se
bioinfo.se
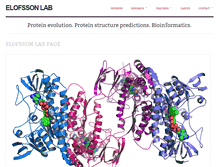
Elofsson Lab – Protein evolution. Protein structure predictions. Bioinformatics.
Master thesis Project Information. Reading instructions for oral exam. Reading material for Classical papers. Protein evolution. Protein structure predictions. Bioinformatics.
 bioinfo.sookmyung.ac.kr
bioinfo.sookmyung.ac.kr
Lab of Bioinformatics and Molecular Design
연도별 저널분류 [SCI,SCIE, JCR]와 Impact Factor확인. Mutation-oriented analysis of che- mical response in cancer cell lines. A novel gene-specific analysis of microarray global data. Prediction of chameleon seq. and amyloid fibril formation. Analysis of geometric arrangement. Smart screening for systems medicines. For this purpose, we have constructed a smart screening platform combining technologies on computer-oriented big data mining and experimental high content screening for last several years. System-level i...
 bioinfo.ssu.ac.kr
bioinfo.ssu.ac.kr
의생명시스탬학부
자동등록방지를 위해 보안절차를 거치고 있습니다. Please prove that you are human.
 bioinfo.swu.bg
bioinfo.swu.bg
ЦЕНТЪР ЗА СЪВРЕМЕННИ БИОИНФОРМАТИЧНИ ИЗСЛЕДВАНИЯ
ЦЕНТЪР ЗА СЪВРЕМЕННИ БИОИНФОРМАТИЧНИ ИЗСЛЕДВАНИЯ. CENTER FOR ADVANCED BIOINFORMATICS RESEARCH. Центърът за Съвременни Биоинформатични Изследвания (ЦСБИ) при ЮЗУ Неофит Рилски , Благоевград е обособена университетска структура за осъществяване на:. Научно-изследователска и проектна дейност на преподаватели в областта на:. Практическо обучение, експериментални разработки и проектна дейност на студенти от различни магистърски, бакалавърски и докторантски специалности като:. Осигуряване на условия за практич...
 bioinfo.t.soka.ac.jp
bioinfo.t.soka.ac.jp
Bioinformatics @ Soka University | 創価大学 工学部 生命情報工学科
Mt Fuji from Soka Univ.
 bioinfo.thep.lu.se
bioinfo.thep.lu.se
Lund SCIBLU Bioinformatics Centre
Lund SCIBLU Bioinformatics Centre. We maintain an in-house database repository of a few common and specialised databases accessible through web interfaces and, in some cases, through command line access. Support for projects with need of web services or database connections. Creation and maintainance of selected software applications. Computational Biology & Biological Physics. Lund University, Sölvegatan 14A. SE - 223 62 Lund, Sweden. Phone: 46-46-222 93 47, Fax: 46-46-222 96 86.